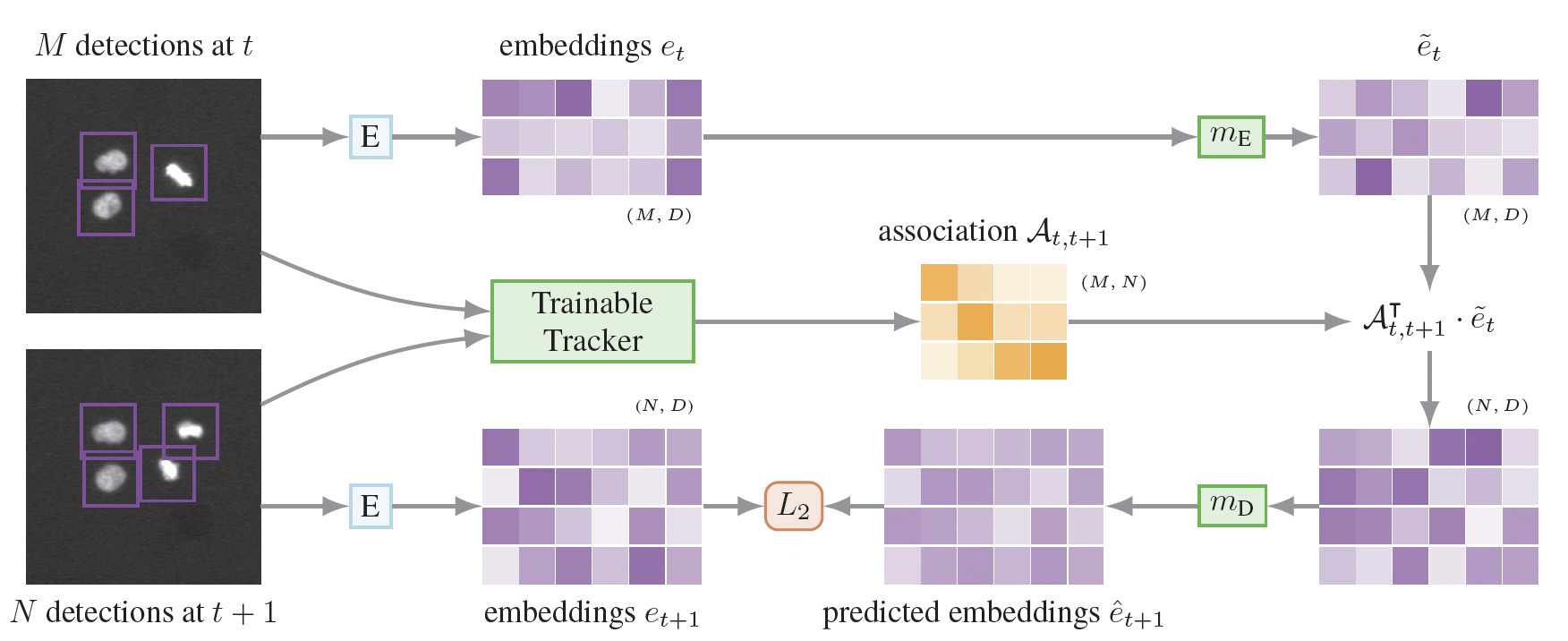
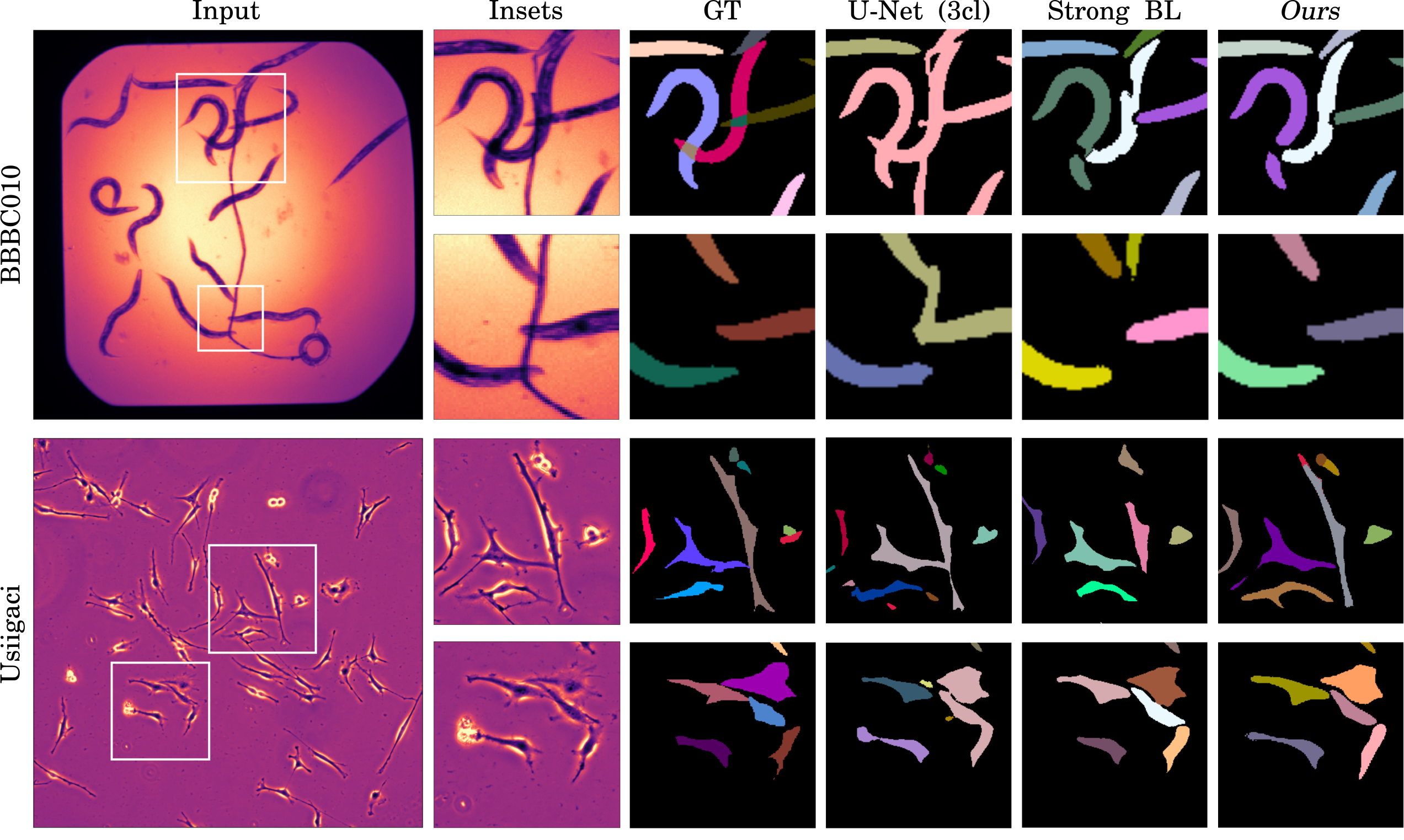An Investigation of Unsupervised Cell Tracking and Interactive Fine-Tuning.
Bio-Image Computing Workshop. International Conference on Computer Vision (ICCV), 2025.
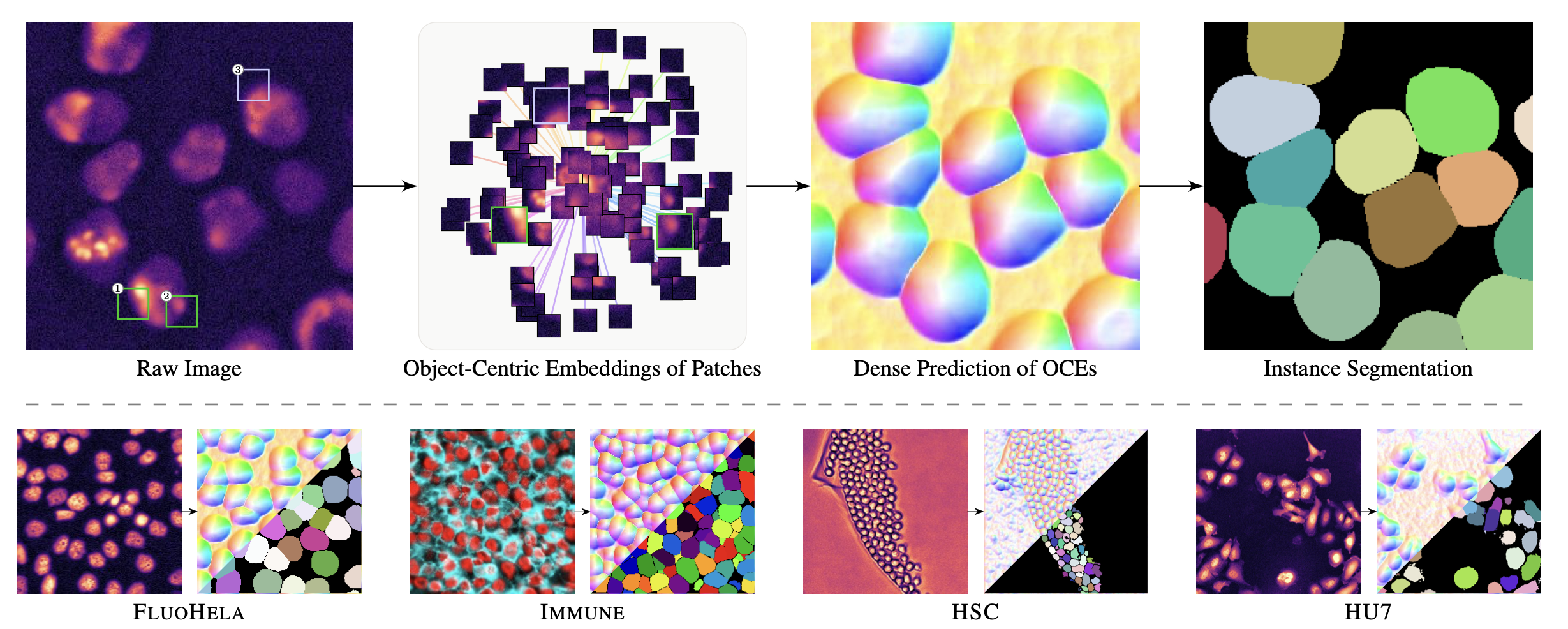
Unsupervised Learning of Object-Centric Embeddings for Cell Instance Segmentation in Microscopy Images.
International Conference on Computer Vision (ICCV), 2023.
DOI: 10.48550/arXiv.2310.08501
Nanog organizes Transcription Bodies.
Current Biology, 2023.
DOI: 10.1016/j.cub.2022.11.015
EmbedSeg: Embedding-based Instance Segmentation for Biomedical Microscopy Data.
Medical Image Analysis, 2022.
DOI: 10.1016/j.media.2022.102523
Embedding-based Instance Segmentation in Microscopy.
Medical Imaging with Deep Learning (MIDL), 2021.
DOI: proceedings.mlr.press/v143/lalit21.html
Modeling expression ranks for noise-tolerant differential expression analysis of scRNA-seq data.
Genome Research, 2021.
DOI: 10.1101/gr.267070.120
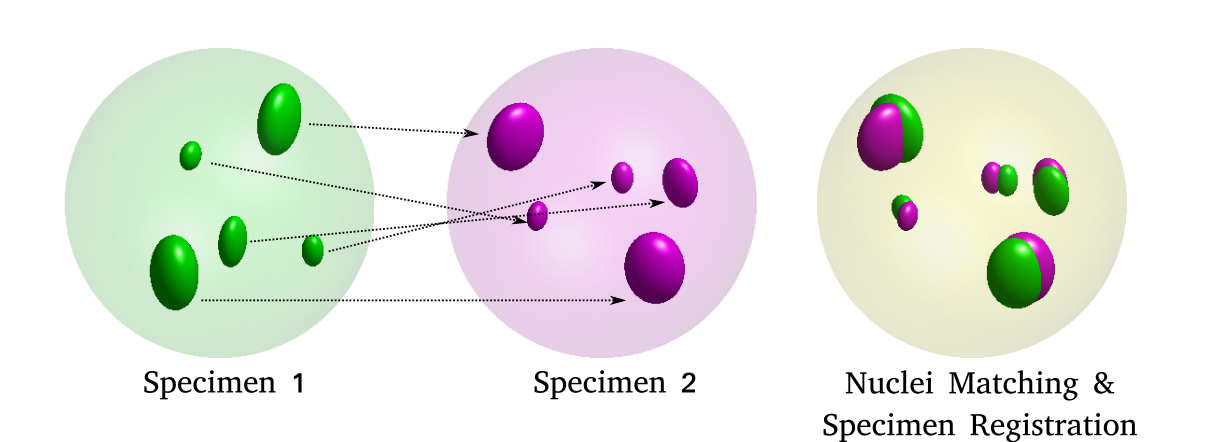
Registration of Multi‐modal Volumetric Images by Establishing Cell Correspondence.
Bio-Image Computing Workshop. European Conference on Computer Vision (ECCV), 2020.
DOI: 10.1007/978-3-030-66415-2_30
Fully Unsupervised Probabilistic Noise2Void.
International Symposium on Biomedical Imaging (ISBI), 2020.
DOI: 10.48550/arXiv.1911.12291
Leveraging Self-supervised Denoising for Image Segmentation.
International Symposium on Biomedical Imaging (ISBI), 2020.
DOI: 10.48550/arXiv.1911.12239
Probabilistic Noise2Void: Unsupervised Content-Aware Denoising.
Frontiers in Computer Science, 2020.
DOI: 10.3389/fcomp.2020.00005
A Note on the Applicability of Thermo-Acoustic Engines for Automotive Waste Heat Recovery.
SAE International Journal of Materials and Manufacturing, 2016.
DOI: 10.4271/2016-01-0223